Constructing a basic data package in Python
As a machine learning researcher, I am constantly scraping, merging, reshaping, exploring, modeling, and generating data. Because I do most of my data management and analysis in Python, I find it convenient to package my data in Python as well. The screenshots below are a walk-through of basic data package construction in Python. In this case, I was programming in a Jupyter notebook and handling a small dataset that I scraped from a protein informatics database. The original Python script can be accessed on GitHub.
The dataset that I am working with describes human peptidase families, as scraped from the MEROPS Peptidase Database in June 2020.
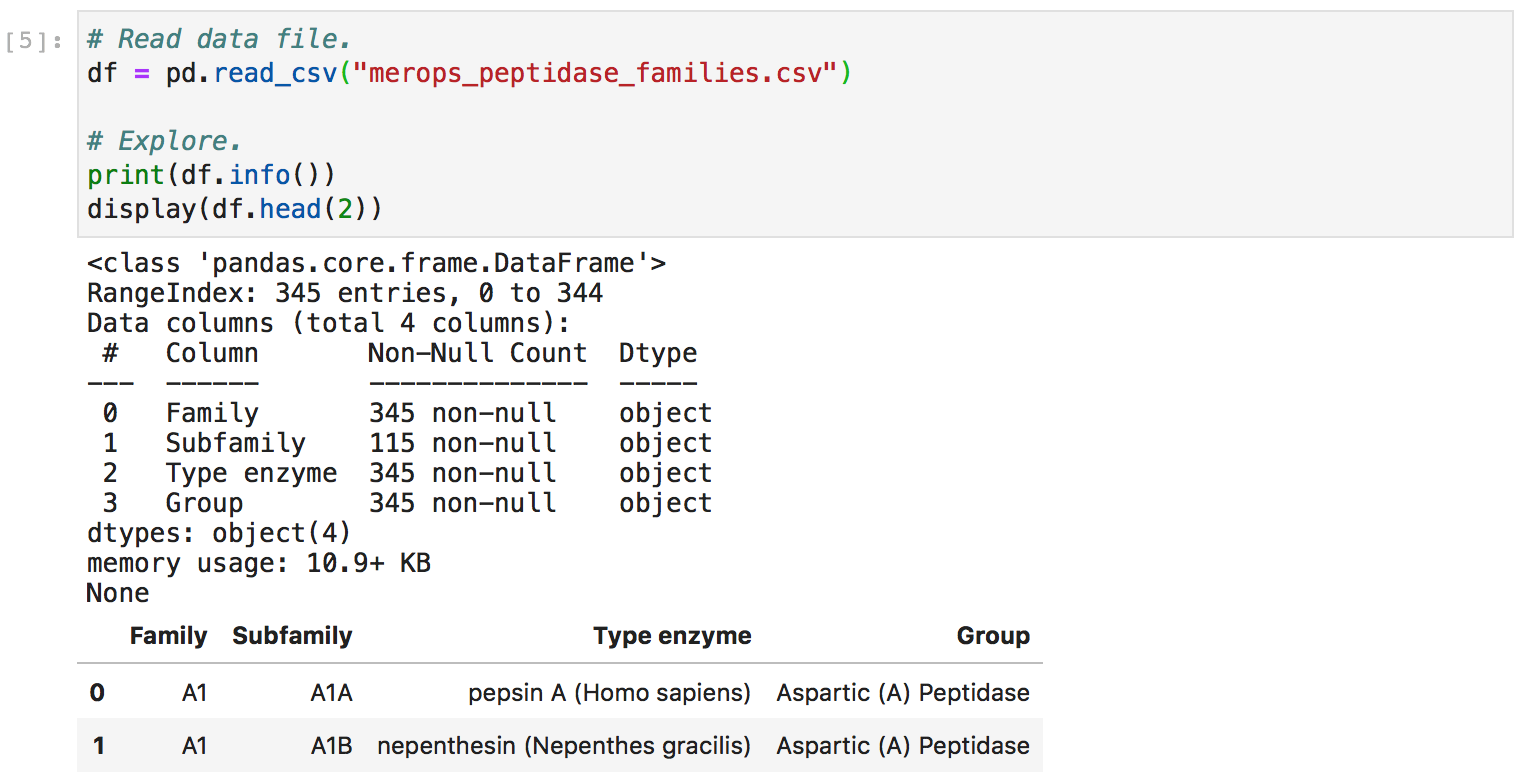
First, I invoked the infer() method to autopopulate the majority of the fields in my data package. I also tested whether my inferred package was valid.
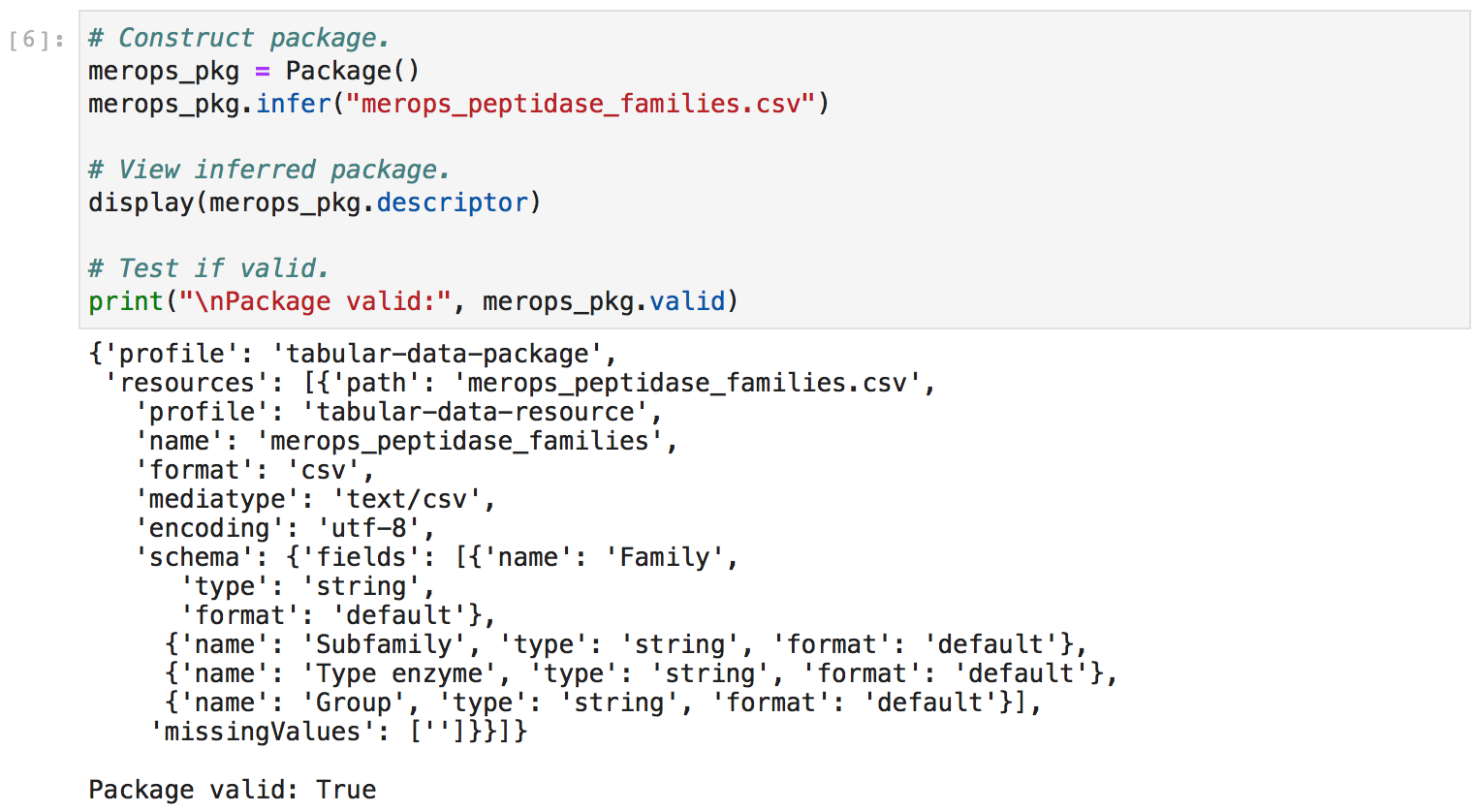
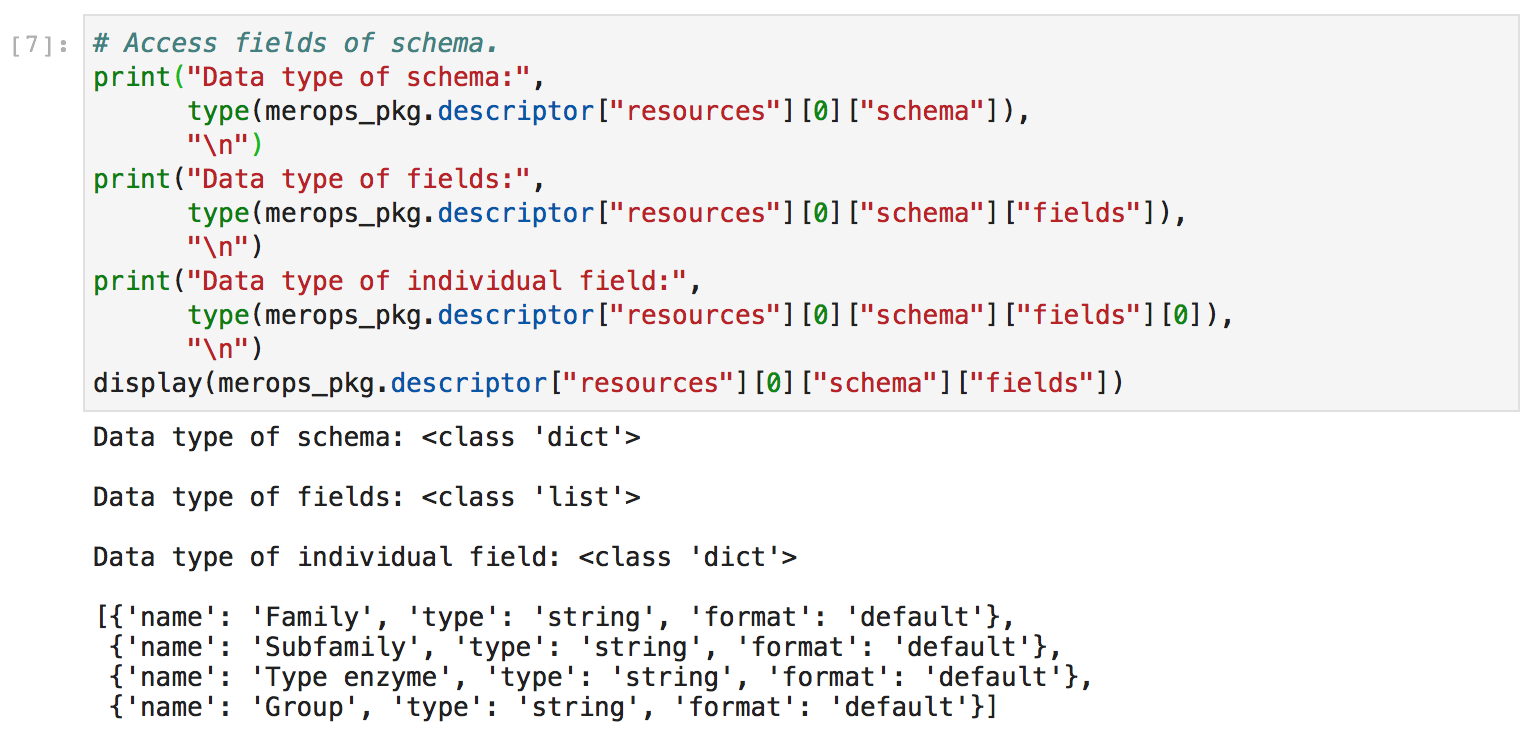
Second, I explored the data structures nested within the merops_pkg.descriptor object. This gave me a better sense of how to manually manipulate these data structures.
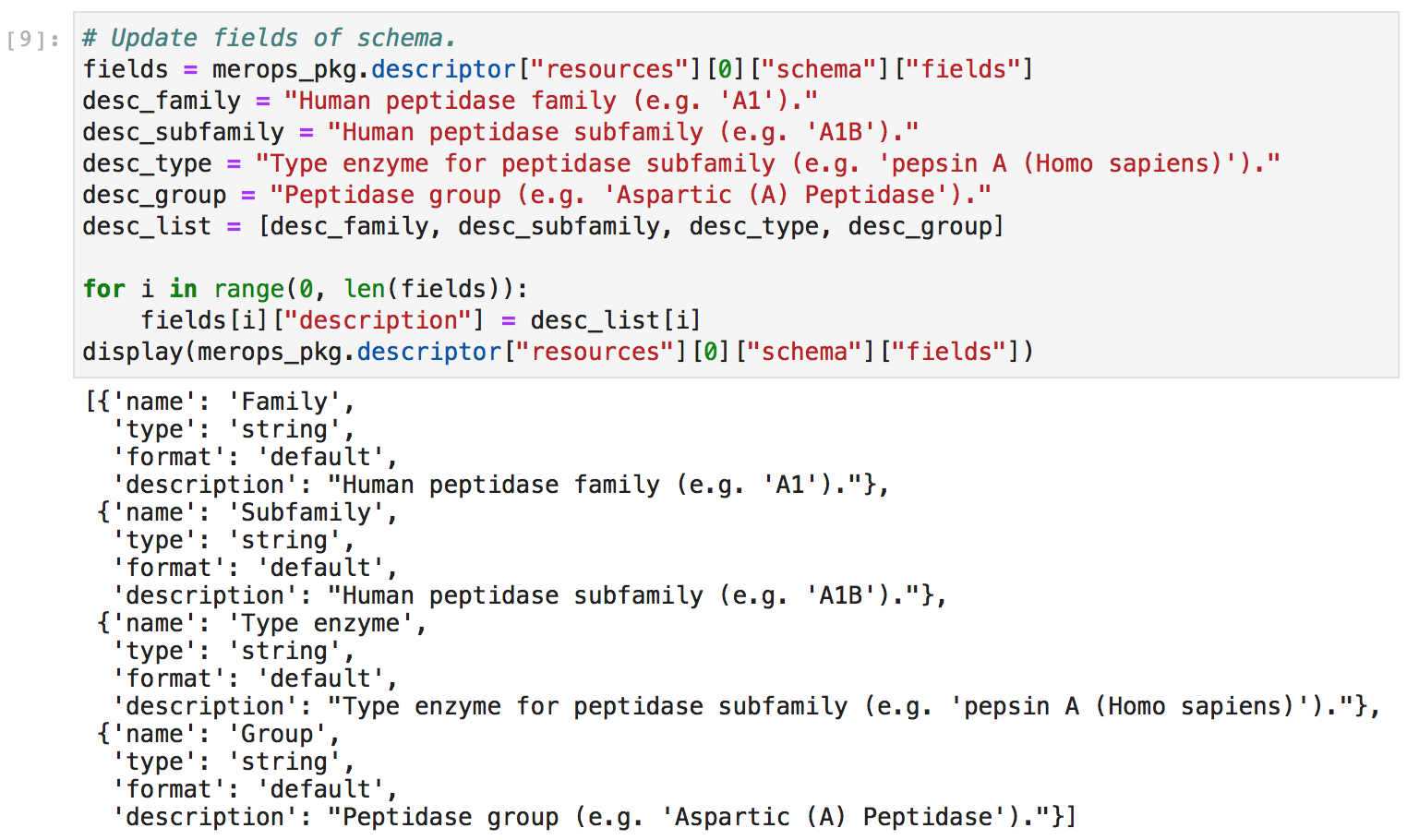
Next, I manually updated the metadata to provide more ample descriptions of the columns in my CSV file. Headings are simply not enough! I want to ensure that even a stranger could easily discern the meaning of each column.
I also added licensing, authorship, keyword, title, and package description metadata to make my data package more useful and informative.
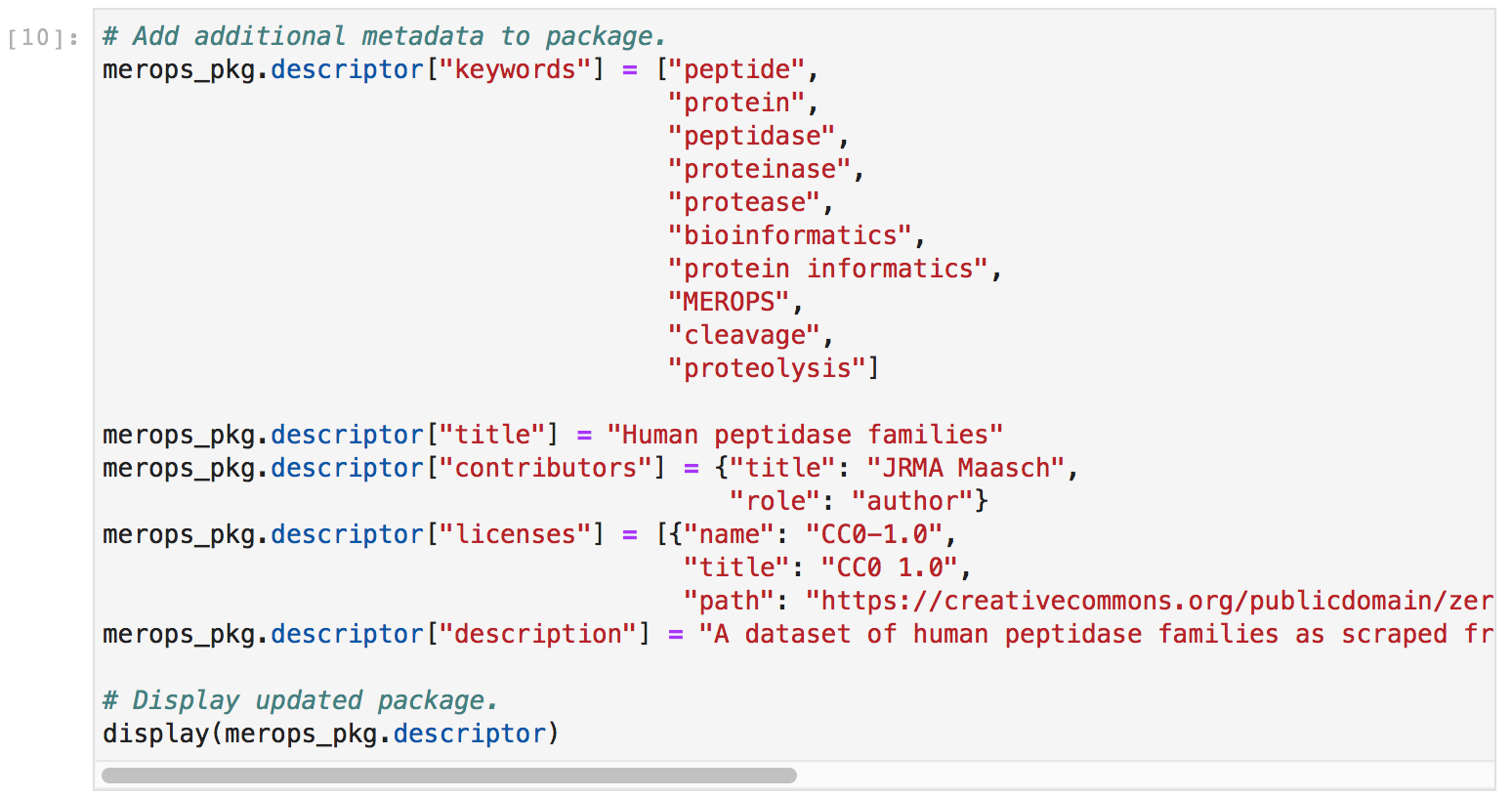
Finally, I committed my changes and saved the data package as a zip file. This zip file contains both the original CSV file and the metadata as a JSON data package file, neatly bundled together.
In the end, my JSON file looks like this:
{
"profile": "tabular-data-package",
"resources": [
{
"path": "data/merops_peptidase_families.csv",
"profile": "tabular-data-resource",
"name": "merops_peptidase_families",
"format": "csv",
"mediatype": "text/csv",
"encoding": "utf-8",
"schema": {
"fields": [
{
"name": "Family",
"type": "string",
"format": "default",
"description": "Human peptidase family (e.g. 'A1')."
},
{
"name": "Subfamily",
"type": "string",
"format": "default",
"description": "Human peptidase subfamily (e.g. 'A1B')."
},
{
"name": "Type enzyme",
"type": "string",
"format": "default",
"description": "Type enzyme for peptidase subfamily (e.g. 'pepsin A (Homo sapiens)')."
},
{
"name": "Group",
"type": "string",
"format": "default",
"description": "Peptidase group (e.g. 'Aspartic (A) Peptidase')."
}
],
"missingValues": [
""
]
}
}
],
"keywords": [
"peptide",
"protein",
"peptidase",
"proteinase",
"protease",
"bioinformatics",
"protein informatics",
"MEROPS",
"cleavage",
"proteolysis"
],
"title": "Human peptidase families",
"contributors": {
"title": "JRMA Maasch",
"role": "author"
},
"licenses": [
{
"name": "CC0-1.0",
"title": "CC0 1.0",
"path": "https://creativecommons.org/publicdomain/zero/1.0/"
}
],
"description": "A dataset of human peptidase families as scraped from the MEROPS Peptidase
Database in June 2020 (https://www.ebi.ac.uk/merops/index.shtml)."
}
This semi-automated process keeps the jungle of data that I obtain, analyze, and produce as orderly and well-documented as possible. Not only does that keep me organized, but it also makes sharing my data frictionless, responsible, and easy.