Validating your data before sharing with the community
In a previous blog post we learnt how to create a Data Package to share the data from your scientific publications.
But often times we need to update or modify our data after we cleaned and share them (e.g., after collecting new observations, or correcting for some errors). This can lead to some errors, such as entering duplicate rows by mistake or using a different format that does not match the one specified before. To avoid these situations, we have a Frictionless tool that helps us verify and validate our dataset before sharing it: goodtables.
As with other Frictionless tools, we can use a GUI-based web application to click our way through the validation process. But this time we will be using the command-line tools and Python library to see how they offer much more flexibility.
Since we uploaded both the CSV file with the data and the JSON file with the metadata to a GitHub repository, we will clone it to work with it locally in our computer:
git clone https://github.com/danalclop/Alcala-Lopez18CerebCortex.git
Once we have the frictionless tools installed (you can check the installation guide for some help if you don't), we can simply get inside the folder we just cloned and use the command line to call goodtables:
goodtables datapackage.json
This will display a bunch of information, starting with a short description of the dataset which will point out whether, overall, the Data Package is valid. As you can see in the image below, the Data Package we created in the previous blog post is valid. But what would had happen if the dataset contained any errors?
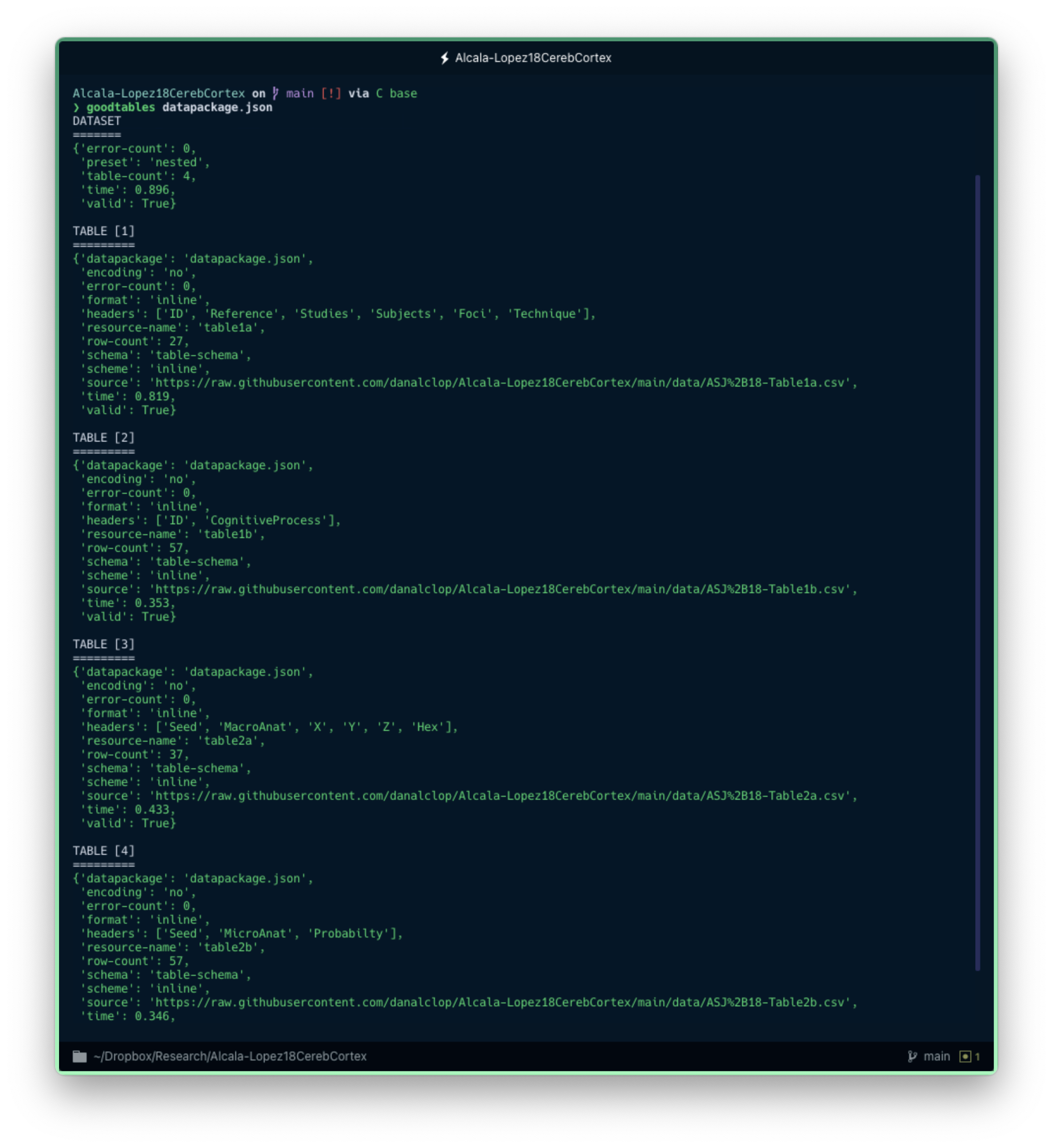
The command-line tool can quickly give you a verification check on your Data Package.
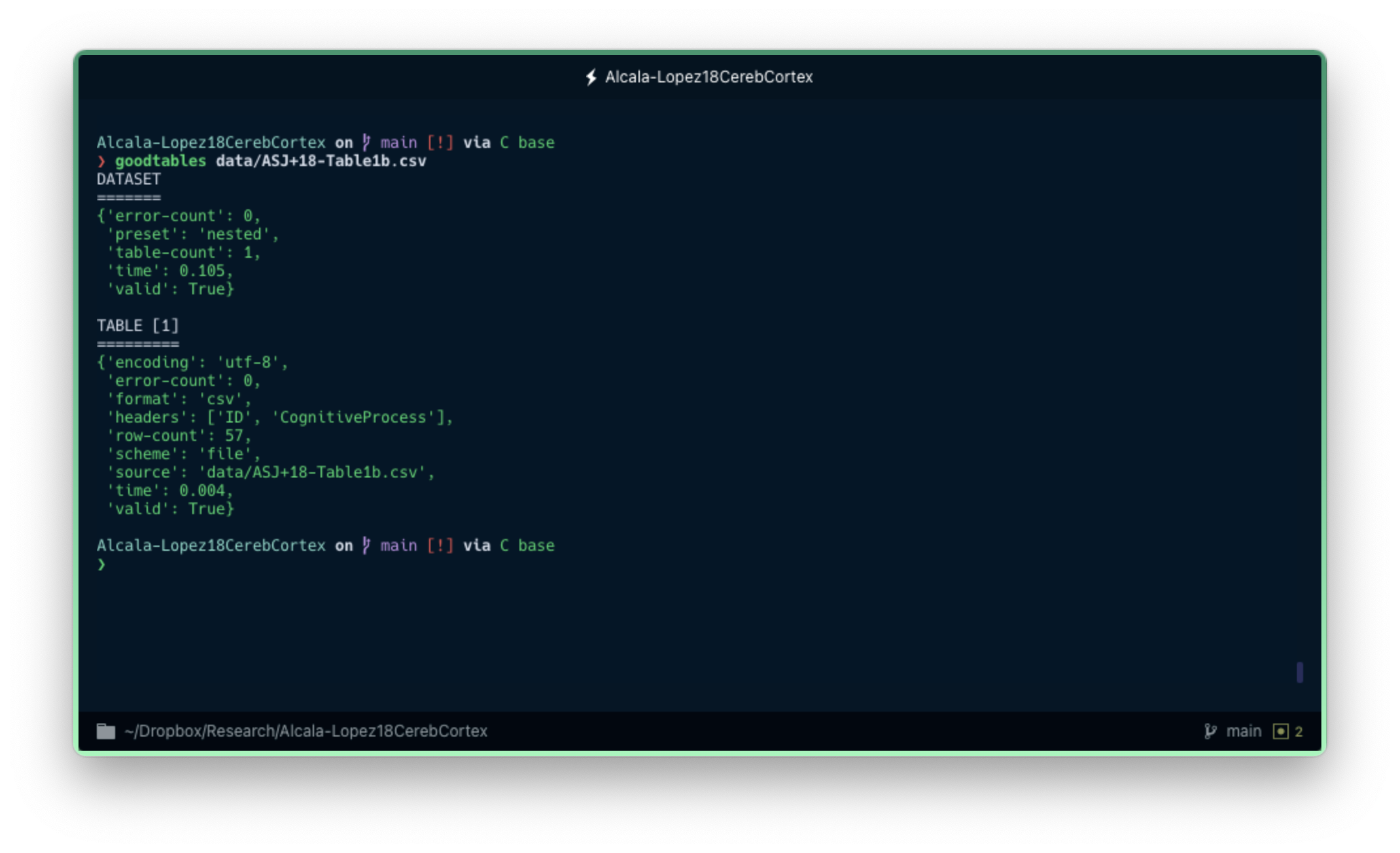
I introduced a few structure errors in a copy of one of the CSV files. Let's see what happens now when we call goodtables from the command line to validate both the previous and the new versions of that data file specifically (instead of the whole Data Package, for simplicity):
goodtables data/ASJ+18-Table1b.csv
goodtables data/ASJ+18-Table1b_invalid.csv
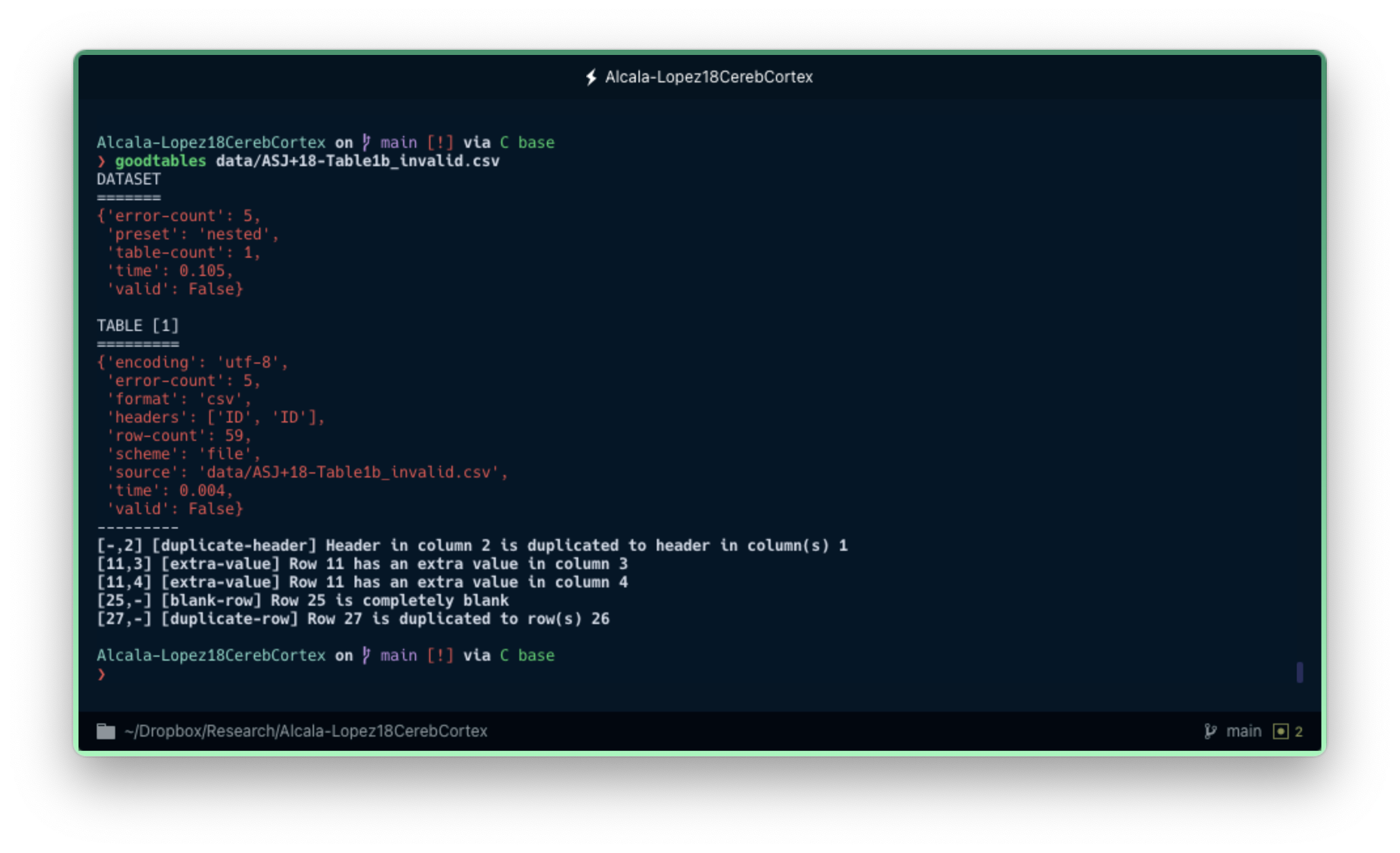
When Goodtables detects an error, it will display a red error message and explain at the bottom what isn't working.
As expected, Goodtables says that the data is not valid. The tool found all the errors and pointed them out so that we can know specifically what each error is and where it is located (row and column). We have a duplicate variable name in the header ('ID' appears twice), a row that contains two extra fields compared to the rest of the data, a row that is entirely blank, and a duplicate row. You can check the list of data quality errors that Goodtables can detect by default, but also create your own custom checks.
This way, you can prevent unseen mistakes or unwanted changes that may affect how re-usable your data is by validating before sharing.
In addition to the command-line interface, we can use the Python library to address each of the errors in detail. This can be helpful if you are already working with your data in Python and want to interactively check the quality of the data:
> from goodtables import validate
> report = validate('data/ASJ+18-Table1b_invalid.csv')
> report['valid']
False
> report['error-count']
5
# print the code of the first error
> report['tables'][0]['errors'][0]['code']
'duplicate-header'
# print the error message of the fourth error
> report['tables'][0]['errors'][3]['message']
'Row 25 is completely blank'
Moreover, there are other things you can do with the Goodtables Python library. You can create your own custom checks or skip one of the existing if, for instance, you deliberately left an entire row blank:
> report = validate('data/ASJ+18-Table1b_invalid.csv', skip_checks=['blank-row', 'missing-value'])
4
Conclusion
Once we have decided to share our data with the rest of the world, it is important to make sure that other people will be able to reuse it. This means providing as much metadata as possible, but also checking that there are no errors in the data that might prevent others from benefiting from our data. Goodtables is a simple tool that you can use both on the web and in the command-line interface to carry out this verification process.
Bonus
If you are planning to share your data and metadata in GitHub, there is a Goodtables website that offers a continuous validation service for tabular data. This means you can sync your GitHub repository and rely on Goodtables to validate your data after every change. You can even add a Goodtables badge at the top of your README file so everyone can see you validated your data before sharing!
Goodtables integrates well with GitHub, offering a continuous validation service for tabular data.