Validate it the GoodTables way!
A basic table is made up of rows and columns with respective data filling up each available cell. Errors may sometimes occur while describing data in a tabular format and these could be in the structure; such as missing headers and duplicated rows, or in the content for instance assigning the wrong character to a string. Some of these errors could be easily spotted by naked eyes and fixed during the data curation process while others may just go unnoticed and later impede some downstream analytical workflows. GoodTables are handy in flagging down common errors that come with tabular data handling as it recognises these discrepancies fast and efficiently to enable users debug their data easily. Validating data files on goodtables therefore acts as the green light for the next phase of data manipulation.There are two ways to validate data: the one time validation in which data files are validated in a single process and the continuous data validation is automated for time to time validation.
One-time data validation
This can be executed in either of two pipelines: the web-based tool and the Command Line Interface (CLI). This blog is about how I used both pipelines to validate my data files (.json and .csv) to check for structural errors. I used this csv file that contains insect counts that I documented from my field visit in smallholder avocado farms of Murang’a, Kenya and a data package that I created in my previous blog. Let's do this!
Try.goodtables.io for Validation
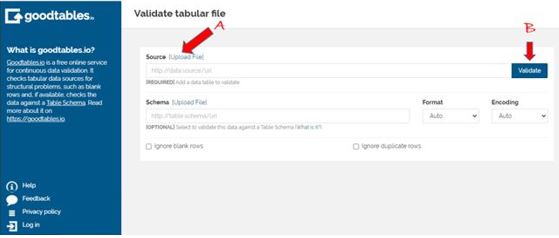
The goodtables web interface
I checked out section A and it provided me with the options for uploading data and I chose to do so from my local computer. After the file was loaded I clicked the validate (B) section and this is what I got:
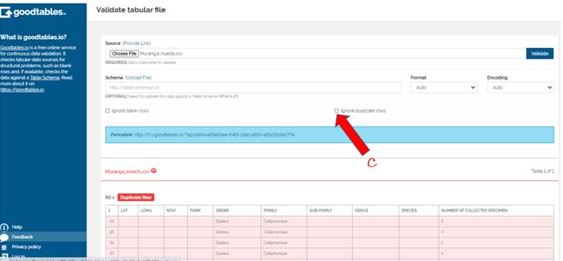
Invalid file with multiple duplicate rows
The program found multiple duplicate rows in my data. This did not worry me though because that’s how I intended those particular fields to appear. To remove these errors, I just clicked the part C (Ignore blank rows) and hit validate again:
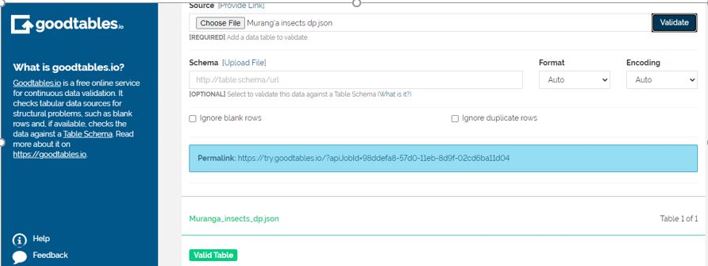
My table is valid!
Ta-da!
While it was simple for me to ignore the duplicate rows, other structural errors such as blank headers may need to be rechecked before repeating the process.
Validating a data package follows the same procedures and if the program detects an error then that will be highlighted promptly and be displayed as above.
Validating using the Command Line Interface (CLI)
For starters, you need these installed:
Python, the programming language in which goodtables is written (latest version) and
PIP, a package installer for python
Basic knowledge on how to use the command line is also a necessity, to refresh on this knowledge see here
First, I installed goodtables on the CLI using pip as the package manager:

Next, I navigated to my working directory using the cd function and once there I invoked the goodtables validate command with the respective file path. First I validated my data package as illustrated below:
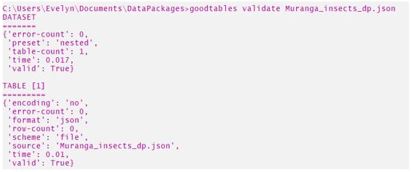
Validated my data package
This was quite a simple process but because I wanted to see how the CLI highlights errors, I chose a csv file containing the same information as the data package and this is how it went:

oops!
This was interesting because both file formats contained the same information, one got validated while the other did not. When using the web interface I had the option of ignoring duplicated rows while the command line did not provide that. I am sure though that there are a set of commands for fixing this. As shown above, the CLI was able to outline the structural errors that dominated my data file in a fraction of a minute.
Starting this exercise was a little technical for me since I forgot to establish my working directory and constantly got an error message that shouted:
[-,-] [scheme-error] [Errno 2] No such file or directory: 'Muranga_insects_dp.json'
So, establishing your working directory is a critical step when working on the commandline (note to self)

I loved this experience because not only was it new, but also I got my results instantly. I validated a lot of data files after this just to make the thrill last longer . I highly recommend goodtables to anyone who’s working with data, if not then get some dummy data and just see for yourself!.