Validating data from Daniel Alcalá-López
In a fast paced research world where there’s an approximate increase of 8-9% in scientific publications every year, an overload of information is usually fed to the outside world. Unfortunately for us, most of this information is often wasted due to the reproducibility crisis marred by data or code that’s often locked away. We explored the question, ‘how reproducible is your data?’ by exchanging personal data and validating them according to the instructions that are outlined in the fellows’ recent goodtables blogs.
Dani’s data
In his paper , Daniel Alcalá-López used brain-imaging techniques to construct a comprehensive neural map of the ‘social brain’.

He used some of the data from this paper in his previous blog and I’m going to do the same and assess how much it takes to reproduce his work and if there’s any challenge along the way. Dani had previously validated his data package and two csv files that he made available on his github account using goodtables. He used the github commandline tool and Python library for this. I replicated his work using the commandline tool by first cloning his repository into my local computer:
$ git clone https://github.com/danalclop/Alcala-Lopez18CerebCortex
With a simplified cd into the folder, I was able to instruct goodtables to validate the datapackage by:
$ goodtables datapackage.json
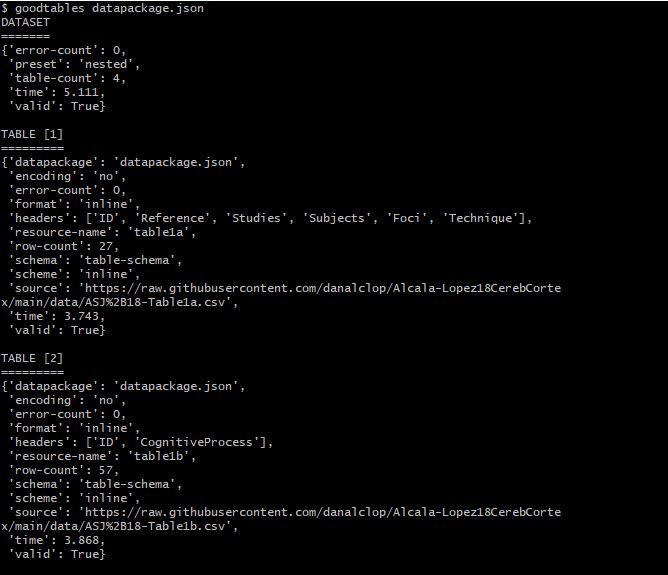
The data package has been validated
Next, I validated a csv file in the same folder by:
$ goodtables data/ASJ+18-Table1b.csv
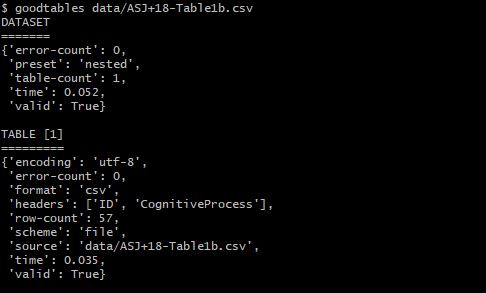
The csv table is also valid
From his last blog, Dani went ahead to introduce some errors to the file above and saved it in a new file for the purposes of testing how goodtables would treat such an obviously invalid file. This I also replicated as shown:
$ goodtables data/ASJ+18-Table1b_invalid.csv
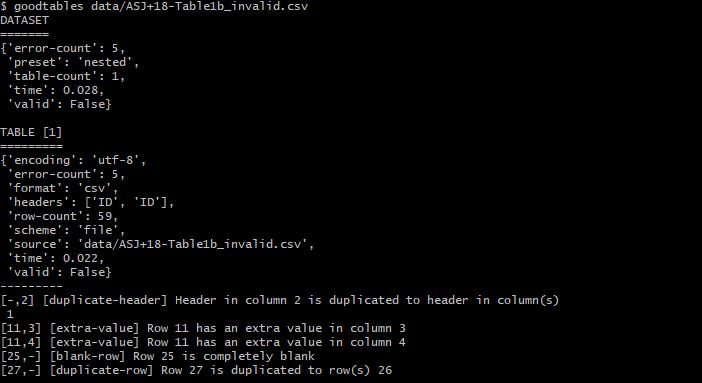
Aha! Goodtables found the errors and outlined them, the csv file therefore is invalid!
I also validated these files using the Python library and what intrigued me most was the overall simplicity of these commands and the detailed nature of error assessment. Also, I give credit to Dani whose blog was self- explanatory and easy to follow.
Additionally...
While conducting this exercise, Dani and I had a zoom meeting where he demonstrated how to conduct continuous data validation using goodtables in github using his data. I just clicked the http://goodtables.io/ url and integrated the service with my github account. The application immediately sifted through my publicly available data and look what I found:
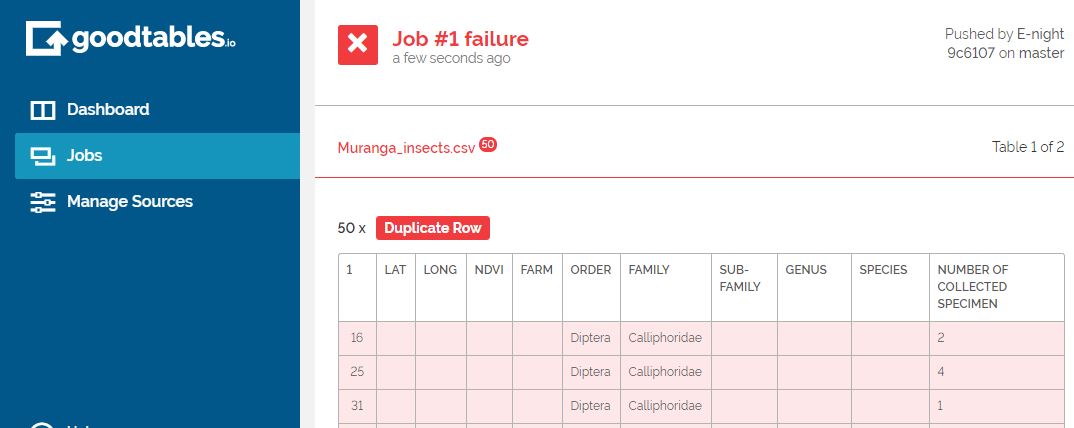
Amazing!
I can now rectify the errors or can even request my collaborators to do so with so much ease. If I upload more files onto my github page then goodtables will automatically validate the files for me!
This exercise was refreshing, to say the least. It was really exciting getting to copy my colleague’s work and get the same results without hustling too much. Ideally this is how scientific data and code ought to be, if I could access and validate brain-imaging data so frictionlessly then I am convinced that it would be easier for researchers from the same field to reproduce each other’s work thereby fast-tracking the process of scientific discoveries and innovations.